Lesson 5. Calculate Summary Values Using Spatial Areas of Interest (AOIs) including Shapefiles for Climate Data Variables Stored in NetCDF 4 Format: Work With MACA v2 Climate Data in Python
Chapter 13 - NETCDF 4 Climate Data in Open Source Python
In this chapter, you will learn how to work with Climate Data Sets (MACA v2 for the United states) stored in netcdf 4 format using open source Python.
Learning Objectives
After completing this chapter, you will be able to:
- Summarize MACA v 2 climate data stored in netcdf 4 format by seasons across all time periods using
xarray. - Summarize MACA v 2 climate data stored in netcdf 4 format by seasons and across years using
xarray.
What You Need
You will need a computer with internet access to complete this lesson and the earth-analytics-python conda environment installed.
Subset MACA 2 Climate Data in NetCDF 4 Format By Time and Spatial Extents
In this lesson, you will learn how to subset MACA v2 climate data using the open source Python packages xarray and regionmask.
import os
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import cartopy.crs as ccrs
import cartopy.feature as cfeature
import seaborn as sns
import geopandas as gpd
import earthpy as et
import xarray as xr
# Spatial subsetting of netcdf files
import regionmask
# Plotting options
sns.set(font_scale=1.3)
sns.set_style("white")
# Optional - set your working directory if you wish to use the data
# accessed lower down in this notebook (the USA state boundary data)
os.chdir(os.path.join(et.io.HOME, 'earth-analytics', 'data'))
Spatial Subsets of Data Using an AOI
In the previous lesson, you learned how to select a single point and extract temperature data at that location. You can also create areas of interest (AOIs) that define the geographic region that you wish to extract data for.
Below you will learn how to use a shapefile that contains a boundary area of interest that you wish to subset data for. You will then learn how to subset the data using the
- geographic boundary extent of the AOI (a rectangular extent) and
- using the actual shape boundary of the AOI (example: the state of California)
To begin, you will open up a new netcdf file. This file contains future projected maximum temperature data for the Continental United States(CONUS).
# Get netcdf file
data_path_monthly = 'http://thredds.northwestknowledge.net:8080/thredds/dodsC/agg_macav2metdata_tasmax_BNU-ESM_r1i1p1_rcp45_2006_2099_CONUS_monthly.nc'
# Open up the data
with xr.open_dataset(data_path_monthly) as file_nc:
monthly_forecast_temp_xr = file_nc
# View xarray object
monthly_forecast_temp_xr
<xarray.Dataset>
Dimensions: (lat: 585, crs: 1, lon: 1386, time: 1128)
Coordinates:
* lat (lat) float64 25.06 25.1 25.15 25.19 ... 49.31 49.35 49.4
* crs (crs) int32 1
* lon (lon) float64 235.2 235.3 235.3 235.4 ... 292.9 292.9 292.9
* time (time) object 2006-01-15 00:00:00 ... 2099-12-15 00:00:00
Data variables:
air_temperature (time, lat, lon) float32 ...
Attributes: (12/46)
description: Multivariate Adaptive Constructed Analog...
id: MACAv2-METDATA
naming_authority: edu.uidaho.reacch
Metadata_Conventions: Unidata Dataset Discovery v1.0
Metadata_Link:
cdm_data_type: FLOAT
... ...
contributor_role: Postdoctoral Fellow
publisher_name: REACCH
publisher_email: reacch@uidaho.edu
publisher_url: http://www.reacchpna.org/
license: Creative Commons CC0 1.0 Universal Dedic...
coordinate_system: WGS84,EPSG:4326- lat: 585
- crs: 1
- lon: 1386
- time: 1128
- lat(lat)float6425.06 25.1 25.15 ... 49.35 49.4
- long_name :
- latitude
- standard_name :
- latitude
- units :
- degrees_north
- axis :
- Y
- description :
- Latitude of the center of the grid cell
array([25.063078, 25.104744, 25.14641 , ..., 49.312691, 49.354359, 49.396023])
- crs(crs)int321
- grid_mapping_name :
- latitude_longitude
- longitude_of_prime_meridian :
- 0.0
- semi_major_axis :
- 6378137.0
- inverse_flattening :
- 298.257223563
array([1], dtype=int32)
- lon(lon)float64235.2 235.3 235.3 ... 292.9 292.9
- units :
- degrees_east
- axis :
- X
- description :
- Longitude of the center of the grid cell
- long_name :
- longitude
- standard_name :
- longitude
array([235.227844, 235.269501, 235.311157, ..., 292.851929, 292.893585, 292.935242]) - time(time)object2006-01-15 00:00:00 ... 2099-12-...
- description :
- days since 1900-01-01
array([cftime.DatetimeNoLeap(2006, 1, 15, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2006, 2, 15, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2006, 3, 15, 0, 0, 0, 0, has_year_zero=True), ..., cftime.DatetimeNoLeap(2099, 10, 15, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2099, 11, 15, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2099, 12, 15, 0, 0, 0, 0, has_year_zero=True)], dtype=object)
- air_temperature(time, lat, lon)float32...
- long_name :
- Monthly Average of Daily Maximum Near-Surface Air Temperature
- units :
- K
- grid_mapping :
- crs
- standard_name :
- air_temperature
- height :
- 2 m
- cell_methods :
- time: maximum(interval: 24 hours);mean over days
- _ChunkSizes :
- [ 10 44 107]
[914593680 values with dtype=float32]
- description :
- Multivariate Adaptive Constructed Analogs (MACA) method, version 2.3,Dec 2013.
- id :
- MACAv2-METDATA
- naming_authority :
- edu.uidaho.reacch
- Metadata_Conventions :
- Unidata Dataset Discovery v1.0
- Metadata_Link :
- cdm_data_type :
- FLOAT
- title :
- Monthly aggregation of downscaled daily meteorological data of Monthly Average of Daily Maximum Near-Surface Air Temperature from College of Global Change and Earth System Science, Beijing Normal University (BNU-ESM) using the run r1i1p1 of the rcp45 scenario.
- summary :
- This archive contains monthly downscaled meteorological and hydrological projections for the Conterminous United States at 1/24-deg resolution. These monthly values are obtained by aggregating the daily values obtained from the downscaling using the Multivariate Adaptive Constructed Analogs (MACA, Abatzoglou, 2012) statistical downscaling method with the METDATA (Abatzoglou,2013) training dataset. The downscaled meteorological variables are maximum/minimum temperature(tasmax/tasmin), maximum/minimum relative humidity (rhsmax/rhsmin),precipitation amount(pr), downward shortwave solar radiation(rsds), eastward wind(uas), northward wind(vas), and specific humidity(huss). The downscaling is based on the 365-day model outputs from different global climate models (GCMs) from Phase 5 of the Coupled Model Inter-comparison Project (CMIP3) utlizing the historical (1950-2005) and future RCP4.5/8.5(2006-2099) scenarios.
- keywords :
- monthly, precipitation, maximum temperature, minimum temperature, downward shortwave solar radiation, specific humidity, wind velocity, CMIP5, Gridded Meteorological Data
- keywords_vocabulary :
- standard_name_vocabulary :
- CF-1.0
- history :
- No revisions.
- comment :
- geospatial_bounds :
- POLYGON((-124.7722 25.0631,-124.7722 49.3960, -67.0648 49.3960,-67.0648, 25.0631, -124.7722,25.0631))
- geospatial_lat_min :
- 25.0631
- geospatial_lat_max :
- 49.3960
- geospatial_lon_min :
- -124.7722
- geospatial_lon_max :
- -67.0648
- geospatial_lat_units :
- decimal degrees north
- geospatial_lon_units :
- decimal degrees east
- geospatial_lat_resolution :
- 0.0417
- geospatial_lon_resolution :
- 0.0417
- geospatial_vertical_min :
- 0.0
- geospatial_vertical_max :
- 0.0
- geospatial_vertical_resolution :
- 0.0
- geospatial_vertical_positive :
- up
- time_coverage_start :
- 2091-01-01T00:0
- time_coverage_end :
- 2095-12-31T00:00
- time_coverage_duration :
- P5Y
- time_coverage_resolution :
- P1M
- date_created :
- 2014-05-15
- date_modified :
- 2014-05-15
- date_issued :
- 2014-05-15
- creator_name :
- John Abatzoglou
- creator_url :
- http://maca.northwestknowledge.net
- creator_email :
- jabatzoglou@uidaho.edu
- institution :
- University of Idaho
- processing_level :
- GRID
- project :
- contributor_name :
- Katherine C. Hegewisch
- contributor_role :
- Postdoctoral Fellow
- publisher_name :
- REACCH
- publisher_email :
- reacch@uidaho.edu
- publisher_url :
- http://www.reacchpna.org/
- license :
- Creative Commons CC0 1.0 Universal Dedication(http://creativecommons.org/publicdomain/zero/1.0/legalcode)
- coordinate_system :
- WGS84,EPSG:4326
Open A Shapefile to Use as an AOI
Often you want to subset and summarize climate data by specific regions of interest. In the example below, you will open a natural earth layer that contains state and region boundaries. You will extract a state boundary (California) within the United States to use as an AOI.
# Download natural earth data which contains state boundaries to generate AOI
url = (
"https://naturalearth.s3.amazonaws.com/"
"50m_cultural/ne_50m_admin_1_states_provinces_lakes.zip"
)
states_gdf = gpd.read_file(url)
states_gdf.head()
| featurecla | scalerank | adm1_code | diss_me | iso_3166_2 | wikipedia | iso_a2 | adm0_sr | name | name_alt | ... | name_nl | name_pl | name_pt | name_ru | name_sv | name_tr | name_vi | name_zh | ne_id | geometry | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Admin-1 scale rank | 2 | AUS-2651 | 2651 | AU-WA | None | AU | 6 | Western Australia | None | ... | West-Australië | Australia Zachodnia | Austrália Ocidental | Западная Австралия | Western Australia | Batı Avustralya | Tây Úc | 西澳大利亚州 | 1159315805 | MULTIPOLYGON (((113.13181 -25.95199, 113.14823... |
| 1 | Admin-1 scale rank | 2 | AUS-2650 | 2650 | AU-NT | None | AU | 6 | Northern Territory | None | ... | Noordelijk Territorium | Terytorium Północne | Território do Norte | Северная территория | Northern Territory | Kuzey Toprakları | Lãnh thổ Bắc Úc | 北領地 | 1159315809 | MULTIPOLYGON (((129.00196 -25.99901, 129.00196... |
| 2 | Admin-1 scale rank | 2 | AUS-2655 | 2655 | AU-SA | None | AU | 3 | South Australia | None | ... | Zuid-Australië | Australia Południowa | Austrália Meridional | Южная Австралия | South Australia | Güney Avustralya | Nam Úc | 南澳大利亚州 | 1159313267 | MULTIPOLYGON (((129.00196 -31.69266, 129.00196... |
| 3 | Admin-1 scale rank | 2 | AUS-2657 | 2657 | AU-QLD | None | AU | 5 | Queensland | None | ... | Queensland | Queensland | Queensland | Квинсленд | Queensland | Queensland | Queensland | 昆士蘭州 | 1159315807 | MULTIPOLYGON (((138.00196 -25.99901, 138.00174... |
| 4 | Admin-1 scale rank | 2 | AUS-2660 | 2660 | AU-TAS | None | AU | 5 | Tasmania | None | ... | Tasmanië | Tasmania | Tasmânia | Тасмания | Tasmanien | Tasmanya | Tasmania | 塔斯馬尼亞州 | 1159313261 | MULTIPOLYGON (((147.31246 -43.28038, 147.34238... |
5 rows × 84 columns
# You will use the bounds to determine the slice values for this data
# Select any state in the CONUS that you wish here! California is the default
cali_aoi = states_gdf[states_gdf.name == "California"]
# Get the total spatial extent for California
cali_aoi.total_bounds
array([-124.37165376, 32.53336527, -114.12501824, 42.00076797])
Next, convert the bounds of your AOI into the min and max longitude and latitude values. You will use these values to slice your data.
# Get lat min, max
aoi_lat = [float(cali_aoi.total_bounds[1]), float(cali_aoi.total_bounds[3])]
aoi_lon = [float(cali_aoi.total_bounds[0]), float(cali_aoi.total_bounds[2])]
# Notice that the longitude values have negative numbers
# we need these values in a global crs so we can subtract from 360
aoi_lat, aoi_lon
([32.533365269889316, 42.00076797479207],
[-124.3716537616361, -114.12501823892204])
The netcdf files use a global lat/lon rather than positive and negative longitude values. To ensure you are subsetting the data for the correct region, you can add 360 degrees to each longitude value which represent the min x and max x values for the extent.
# The netcdf files use a global lat/lon so adjust values accordingly
aoi_lon[0] = aoi_lon[0] + 360
aoi_lon[1] = aoi_lon[1] + 360
aoi_lon
[235.62834623836392, 245.87498176107795]
Once you have your data AOI defined, you can slice out the AOI region using xarray slice.
# Slice the data by time and spatial extent
start_date = "2010-01-15"
end_date = "2010-02-15"
two_months_cali = monthly_forecast_temp_xr["air_temperature"].sel(
time=slice(start_date, end_date),
lon=slice(aoi_lon[0], aoi_lon[1]),
lat=slice(aoi_lat[0], aoi_lat[1]))
two_months_cali
<xarray.DataArray 'air_temperature' (time: 2, lat: 227, lon: 246)>
[111684 values with dtype=float32]
Coordinates:
* lat (lat) float64 32.56 32.6 32.65 32.69 ... 41.85 41.9 41.94 41.98
* lon (lon) float64 235.6 235.7 235.7 235.8 ... 245.7 245.8 245.8 245.9
* time (time) object 2010-01-15 00:00:00 2010-02-15 00:00:00
Attributes:
long_name: Monthly Average of Daily Maximum Near-Surface Air Tempera...
units: K
grid_mapping: crs
standard_name: air_temperature
height: 2 m
cell_methods: time: maximum(interval: 24 hours);mean over days
_ChunkSizes: [ 10 44 107]- time: 2
- lat: 227
- lon: 246
- ...
[111684 values with dtype=float32]
- lat(lat)float6432.56 32.6 32.65 ... 41.94 41.98
- long_name :
- latitude
- standard_name :
- latitude
- units :
- degrees_north
- axis :
- Y
- description :
- Latitude of the center of the grid cell
array([32.562958, 32.604626, 32.64629 , ..., 41.896141, 41.937809, 41.979473])
- lon(lon)float64235.6 235.7 235.7 ... 245.8 245.9
- units :
- degrees_east
- axis :
- X
- description :
- Longitude of the center of the grid cell
- long_name :
- longitude
- standard_name :
- longitude
array([235.644501, 235.686157, 235.727829, ..., 245.769333, 245.811005, 245.852661]) - time(time)object2010-01-15 00:00:00 2010-02-15 0...
- description :
- days since 1900-01-01
array([cftime.DatetimeNoLeap(2010, 1, 15, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2010, 2, 15, 0, 0, 0, 0, has_year_zero=True)], dtype=object)
- long_name :
- Monthly Average of Daily Maximum Near-Surface Air Temperature
- units :
- K
- grid_mapping :
- crs
- standard_name :
- air_temperature
- height :
- 2 m
- cell_methods :
- time: maximum(interval: 24 hours);mean over days
- _ChunkSizes :
- [ 10 44 107]
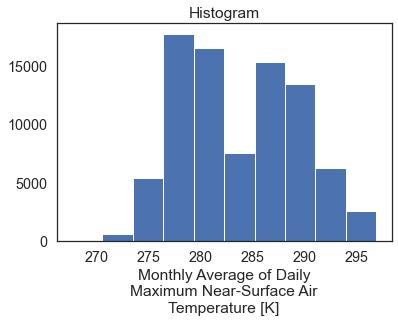
# Plot a quick histogram
two_months_cali.plot()
plt.show()
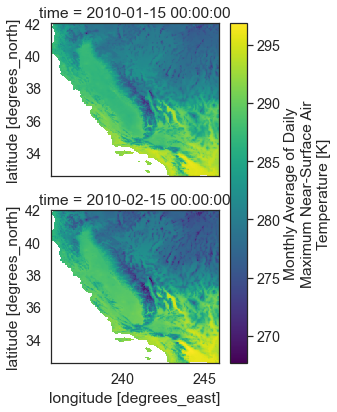
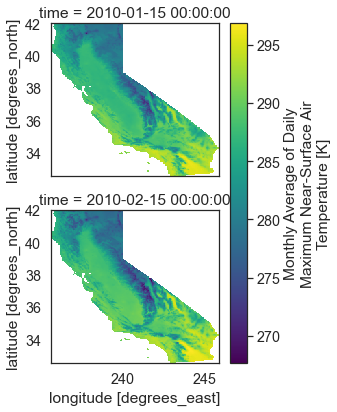
Remember that these data are spatial. Below you plot each time step as a raster dataset. Notice that this spatial extent is a rectangle representing the entire rectangular extent for the state of California.
# Spatial Plot for the selected AOI (California)
two_months_cali.plot(col='time',
col_wrap=1)
plt.show()
# Only subset by location / not time
cali_ts = monthly_forecast_temp_xr["air_temperature"].sel(
lon=slice(aoi_lon[0], aoi_lon[1]),
lat=slice(aoi_lat[0], aoi_lat[1]))
cali_ts
<xarray.DataArray 'air_temperature' (time: 1128, lat: 227, lon: 246)>
[62989776 values with dtype=float32]
Coordinates:
* lat (lat) float64 32.56 32.6 32.65 32.69 ... 41.85 41.9 41.94 41.98
* lon (lon) float64 235.6 235.7 235.7 235.8 ... 245.7 245.8 245.8 245.9
* time (time) object 2006-01-15 00:00:00 ... 2099-12-15 00:00:00
Attributes:
long_name: Monthly Average of Daily Maximum Near-Surface Air Tempera...
units: K
grid_mapping: crs
standard_name: air_temperature
height: 2 m
cell_methods: time: maximum(interval: 24 hours);mean over days
_ChunkSizes: [ 10 44 107]- time: 1128
- lat: 227
- lon: 246
- ...
[62989776 values with dtype=float32]
- lat(lat)float6432.56 32.6 32.65 ... 41.94 41.98
- long_name :
- latitude
- standard_name :
- latitude
- units :
- degrees_north
- axis :
- Y
- description :
- Latitude of the center of the grid cell
array([32.562958, 32.604626, 32.64629 , ..., 41.896141, 41.937809, 41.979473])
- lon(lon)float64235.6 235.7 235.7 ... 245.8 245.9
- units :
- degrees_east
- axis :
- X
- description :
- Longitude of the center of the grid cell
- long_name :
- longitude
- standard_name :
- longitude
array([235.644501, 235.686157, 235.727829, ..., 245.769333, 245.811005, 245.852661]) - time(time)object2006-01-15 00:00:00 ... 2099-12-...
- description :
- days since 1900-01-01
array([cftime.DatetimeNoLeap(2006, 1, 15, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2006, 2, 15, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2006, 3, 15, 0, 0, 0, 0, has_year_zero=True), ..., cftime.DatetimeNoLeap(2099, 10, 15, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2099, 11, 15, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2099, 12, 15, 0, 0, 0, 0, has_year_zero=True)], dtype=object)
- long_name :
- Monthly Average of Daily Maximum Near-Surface Air Temperature
- units :
- K
- grid_mapping :
- crs
- standard_name :
- air_temperature
- height :
- 2 m
- cell_methods :
- time: maximum(interval: 24 hours);mean over days
- _ChunkSizes :
- [ 10 44 107]
Calculate a summary of max temperature over time for california
# Time series plot of max temperature per year. for California
# You will get a RuntimeWarning warning here because of nan values...
# This is the max value in each pixel across all months for each year
cali_annual_max = cali_ts.groupby('time.year').max(skipna=True)
cali_annual_max
<xarray.DataArray 'air_temperature' (year: 94, lat: 227, lon: 246)>
array([[[ nan, nan, nan, ..., 314.2675 , 314.05508,
313.95047],
[ nan, nan, nan, ..., 314.08972, 313.86816,
313.9082 ],
[ nan, nan, nan, ..., 314.02127, 313.8809 ,
313.9658 ],
...,
[ nan, nan, nan, ..., 304.58743, 304.74078,
304.8877 ],
[ nan, nan, nan, ..., 303.80627, 303.83685,
304.6529 ],
[ nan, 293.99442, 294.31528, ..., 302.92322, 302.578 ,
303.59604]],
[[ nan, nan, nan, ..., 314.5754 , 314.44275,
314.34146],
[ nan, nan, nan, ..., 314.50024, 314.32486,
314.3367 ],
[ nan, nan, nan, ..., 314.4508 , 314.33475,
314.3861 ],
...
[ nan, nan, nan, ..., 306.28198, 306.4167 ,
306.61166],
[ nan, nan, nan, ..., 305.46466, 305.49435,
306.3104 ],
[ nan, 296.4974 , 296.71976, ..., 304.55817, 304.22375,
305.2304 ]],
[[ nan, nan, nan, ..., 316.17175, 315.9877 ,
315.91992],
[ nan, nan, nan, ..., 316.05585, 315.89587,
315.90164],
[ nan, nan, nan, ..., 316.0458 , 315.94336,
315.96744],
...,
[ nan, nan, nan, ..., 309.6978 , 310.04214,
310.2721 ],
[ nan, nan, nan, ..., 308.9082 , 308.9424 ,
309.8799 ],
[ nan, 296.43567, 296.677 , ..., 307.94104, 307.59906,
308.6584 ]]], dtype=float32)
Coordinates:
* lat (lat) float64 32.56 32.6 32.65 32.69 ... 41.85 41.9 41.94 41.98
* lon (lon) float64 235.6 235.7 235.7 235.8 ... 245.7 245.8 245.8 245.9
* year (year) int64 2006 2007 2008 2009 2010 ... 2095 2096 2097 2098 2099- year: 94
- lat: 227
- lon: 246
- nan nan nan nan nan nan nan ... 308.3 306.5 307.6 307.9 307.6 308.7
array([[[ nan, nan, nan, ..., 314.2675 , 314.05508, 313.95047], [ nan, nan, nan, ..., 314.08972, 313.86816, 313.9082 ], [ nan, nan, nan, ..., 314.02127, 313.8809 , 313.9658 ], ..., [ nan, nan, nan, ..., 304.58743, 304.74078, 304.8877 ], [ nan, nan, nan, ..., 303.80627, 303.83685, 304.6529 ], [ nan, 293.99442, 294.31528, ..., 302.92322, 302.578 , 303.59604]], [[ nan, nan, nan, ..., 314.5754 , 314.44275, 314.34146], [ nan, nan, nan, ..., 314.50024, 314.32486, 314.3367 ], [ nan, nan, nan, ..., 314.4508 , 314.33475, 314.3861 ], ... [ nan, nan, nan, ..., 306.28198, 306.4167 , 306.61166], [ nan, nan, nan, ..., 305.46466, 305.49435, 306.3104 ], [ nan, 296.4974 , 296.71976, ..., 304.55817, 304.22375, 305.2304 ]], [[ nan, nan, nan, ..., 316.17175, 315.9877 , 315.91992], [ nan, nan, nan, ..., 316.05585, 315.89587, 315.90164], [ nan, nan, nan, ..., 316.0458 , 315.94336, 315.96744], ..., [ nan, nan, nan, ..., 309.6978 , 310.04214, 310.2721 ], [ nan, nan, nan, ..., 308.9082 , 308.9424 , 309.8799 ], [ nan, 296.43567, 296.677 , ..., 307.94104, 307.59906, 308.6584 ]]], dtype=float32) - lat(lat)float6432.56 32.6 32.65 ... 41.94 41.98
- long_name :
- latitude
- standard_name :
- latitude
- units :
- degrees_north
- axis :
- Y
- description :
- Latitude of the center of the grid cell
array([32.562958, 32.604626, 32.64629 , ..., 41.896141, 41.937809, 41.979473])
- lon(lon)float64235.6 235.7 235.7 ... 245.8 245.9
- units :
- degrees_east
- axis :
- X
- description :
- Longitude of the center of the grid cell
- long_name :
- longitude
- standard_name :
- longitude
array([235.644501, 235.686157, 235.727829, ..., 245.769333, 245.811005, 245.852661]) - year(year)int642006 2007 2008 ... 2097 2098 2099
array([2006, 2007, 2008, 2009, 2010, 2011, 2012, 2013, 2014, 2015, 2016, 2017, 2018, 2019, 2020, 2021, 2022, 2023, 2024, 2025, 2026, 2027, 2028, 2029, 2030, 2031, 2032, 2033, 2034, 2035, 2036, 2037, 2038, 2039, 2040, 2041, 2042, 2043, 2044, 2045, 2046, 2047, 2048, 2049, 2050, 2051, 2052, 2053, 2054, 2055, 2056, 2057, 2058, 2059, 2060, 2061, 2062, 2063, 2064, 2065, 2066, 2067, 2068, 2069, 2070, 2071, 2072, 2073, 2074, 2075, 2076, 2077, 2078, 2079, 2080, 2081, 2082, 2083, 2084, 2085, 2086, 2087, 2088, 2089, 2090, 2091, 2092, 2093, 2094, 2095, 2096, 2097, 2098, 2099])
Calculate Annual Summary Data
You can calculate summary values for your aoi using the .groupby method. Below you calculate the max value for each raster in the time series
.max(["lat", "lon"] tells xarray to calculate the max on the entire raster.
the data are grouped by year.
cali_annual_max_val = cali_annual_max.groupby("year").max(["lat", "lon"])
cali_annual_max_val
<xarray.DataArray 'air_temperature' (year: 94)>
array([320.67538, 322.2723 , 320.86505, 321.88 , 321.55954, 320.30484,
322.17026, 322.1134 , 322.16568, 321.32007, 321.67624, 322.07996,
321.5724 , 320.7888 , 321.4106 , 320.6512 , 322.2032 , 322.18158,
322.05664, 322.96497, 322.20325, 321.28458, 321.51407, 324.0671 ,
323.02896, 322.10184, 322.73853, 322.56485, 321.98178, 322.4918 ,
322.19852, 322.44962, 321.97635, 322.33276, 322.55652, 322.4373 ,
322.4933 , 323.7776 , 323.3054 , 321.22037, 322.00476, 322.40897,
323.43805, 323.57452, 322.63904, 321.85776, 324.09442, 323.27408,
323.67053, 322.62167, 323.70203, 322.82205, 321.93048, 323.48462,
323.89087, 323.4939 , 324.15057, 322.88406, 322.6229 , 324.0509 ,
323.06467, 324.27808, 324.30505, 323.27487, 323.3609 , 324.03644,
323.1147 , 324.6987 , 322.52747, 323.00424, 324.0726 , 322.78583,
323.49216, 322.4643 , 321.52664, 323.14383, 323.44632, 323.4093 ,
324.51718, 322.68958, 324.83566, 323.24158, 323.22598, 323.8113 ,
323.70255, 321.93524, 325.1739 , 322.6527 , 323.33313, 323.01407,
323.40854, 323.8004 , 321.59985, 324.6846 ], dtype=float32)
Coordinates:
* year (year) int64 2006 2007 2008 2009 2010 ... 2095 2096 2097 2098 2099- year: 94
- 320.7 322.3 320.9 321.9 321.6 320.3 ... 323.0 323.4 323.8 321.6 324.7
array([320.67538, 322.2723 , 320.86505, 321.88 , 321.55954, 320.30484, 322.17026, 322.1134 , 322.16568, 321.32007, 321.67624, 322.07996, 321.5724 , 320.7888 , 321.4106 , 320.6512 , 322.2032 , 322.18158, 322.05664, 322.96497, 322.20325, 321.28458, 321.51407, 324.0671 , 323.02896, 322.10184, 322.73853, 322.56485, 321.98178, 322.4918 , 322.19852, 322.44962, 321.97635, 322.33276, 322.55652, 322.4373 , 322.4933 , 323.7776 , 323.3054 , 321.22037, 322.00476, 322.40897, 323.43805, 323.57452, 322.63904, 321.85776, 324.09442, 323.27408, 323.67053, 322.62167, 323.70203, 322.82205, 321.93048, 323.48462, 323.89087, 323.4939 , 324.15057, 322.88406, 322.6229 , 324.0509 , 323.06467, 324.27808, 324.30505, 323.27487, 323.3609 , 324.03644, 323.1147 , 324.6987 , 322.52747, 323.00424, 324.0726 , 322.78583, 323.49216, 322.4643 , 321.52664, 323.14383, 323.44632, 323.4093 , 324.51718, 322.68958, 324.83566, 323.24158, 323.22598, 323.8113 , 323.70255, 321.93524, 325.1739 , 322.6527 , 323.33313, 323.01407, 323.40854, 323.8004 , 321.59985, 324.6846 ], dtype=float32) - year(year)int642006 2007 2008 ... 2097 2098 2099
array([2006, 2007, 2008, 2009, 2010, 2011, 2012, 2013, 2014, 2015, 2016, 2017, 2018, 2019, 2020, 2021, 2022, 2023, 2024, 2025, 2026, 2027, 2028, 2029, 2030, 2031, 2032, 2033, 2034, 2035, 2036, 2037, 2038, 2039, 2040, 2041, 2042, 2043, 2044, 2045, 2046, 2047, 2048, 2049, 2050, 2051, 2052, 2053, 2054, 2055, 2056, 2057, 2058, 2059, 2060, 2061, 2062, 2063, 2064, 2065, 2066, 2067, 2068, 2069, 2070, 2071, 2072, 2073, 2074, 2075, 2076, 2077, 2078, 2079, 2080, 2081, 2082, 2083, 2084, 2085, 2086, 2087, 2088, 2089, 2090, 2091, 2092, 2093, 2094, 2095, 2096, 2097, 2098, 2099])
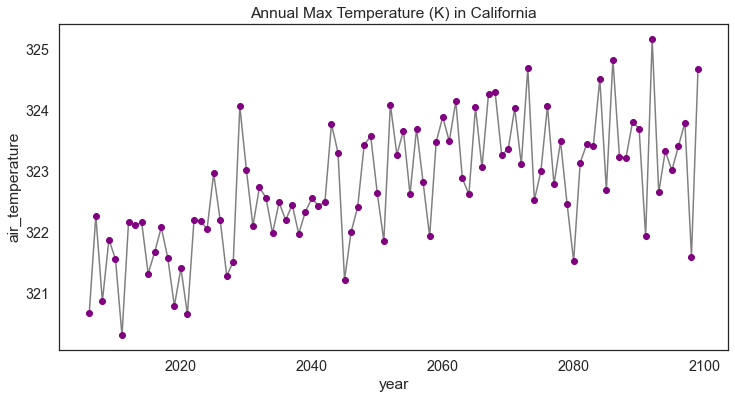
# Plot the data
f, ax = plt.subplots(figsize=(12, 6))
cali_annual_max_val.plot.line(hue='lat',
marker="o",
ax=ax,
color="grey",
markerfacecolor="purple",
markeredgecolor="purple")
ax.set(title="Annual Max Temperature (K) in California")
plt.show()
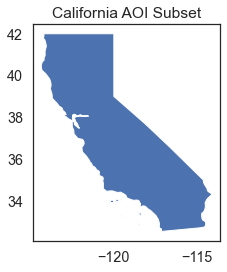
Subset a netcdf4 Using a Shapefile Feature or Features
Above you created a rectangular spatial subset and plotted the data. Sometimes however you may have an AOI that is a specific polygon boundary. Below you will modify your subset to represent just the region of the state of California. You will use the regionmask package to create this mask.
# This is the AOI of interest. You only want to calculate summary values for
# pixels within this AOI rather the entire rectangular spatial extent.
f, ax = plt.subplots()
cali_aoi.plot(ax=ax)
ax.set(title="California AOI Subset")
plt.show()
You will use the cali_aoi object created above to first create a mask.
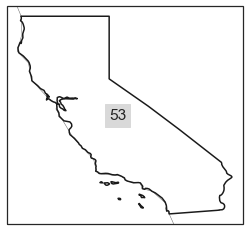
cali_aoi
| featurecla | scalerank | adm1_code | diss_me | iso_3166_2 | wikipedia | iso_a2 | adm0_sr | name | name_alt | ... | name_nl | name_pl | name_pt | name_ru | name_sv | name_tr | name_vi | name_zh | ne_id | geometry | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 53 | Admin-1 scale rank | 2 | USA-3521 | 3521 | US-CA | http://en.wikipedia.org/wiki/California | US | 8 | California | CA|Calif. | ... | Californië | Kalifornia | Califórnia | Калифорния | Kalifornien | Kaliforniya | California | 加利福尼亚州 | 1159308415 | MULTIPOLYGON (((-114.61054 34.99112, -114.6109... |
1 rows × 84 columns
<regionmask.Regions>
Name: name
Regions:
53 US-CA California
[1 regions]
<xarray.DataArray 'region' (lat: 585, lon: 1386)>
array([[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan],
...,
[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan]])
Coordinates:
* lat (lat) float64 25.06 25.1 25.15 25.19 ... 49.27 49.31 49.35 49.4
* lon (lon) float64 235.2 235.3 235.3 235.4 ... 292.8 292.9 292.9 292.9- lat: 585
- lon: 1386
- nan nan nan nan nan nan nan nan ... nan nan nan nan nan nan nan nan
array([[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]) - lat(lat)float6425.06 25.1 25.15 ... 49.35 49.4
array([25.063078, 25.104744, 25.14641 , ..., 49.312691, 49.354359, 49.396023])
- lon(lon)float64235.2 235.3 235.3 ... 292.9 292.9
array([235.227844, 235.269501, 235.311157, ..., 292.851929, 292.893585, 292.935242])
(<xarray.DataArray 'region' ()>
array(53.),
<xarray.DataArray 'region' ()>
array(53.))
<xarray.DataArray 'air_temperature' (time: 2, lat: 227, lon: 246)>
array([[[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan],
...,
[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan]],
[[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan],
...,
[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan]]], dtype=float32)
Coordinates:
* lat (lat) float64 32.56 32.6 32.65 32.69 ... 41.85 41.9 41.94 41.98
* lon (lon) float64 235.6 235.7 235.7 235.8 ... 245.7 245.8 245.8 245.9
* time (time) object 2010-01-15 00:00:00 2010-02-15 00:00:00
Attributes:
long_name: Monthly Average of Daily Maximum Near-Surface Air Tempera...
units: K
grid_mapping: crs
standard_name: air_temperature
height: 2 m
cell_methods: time: maximum(interval: 24 hours);mean over days
_ChunkSizes: [ 10 44 107]- time: 2
- lat: 227
- lon: 246
- nan nan nan nan nan nan nan nan ... nan nan nan nan nan nan nan nan
array([[[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]], [[nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], ..., [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan], [nan, nan, nan, ..., nan, nan, nan]]], dtype=float32) - lat(lat)float6432.56 32.6 32.65 ... 41.94 41.98
- long_name :
- latitude
- standard_name :
- latitude
- units :
- degrees_north
- axis :
- Y
- description :
- Latitude of the center of the grid cell
array([32.562958, 32.604626, 32.64629 , ..., 41.896141, 41.937809, 41.979473])
- lon(lon)float64235.6 235.7 235.7 ... 245.8 245.9
- units :
- degrees_east
- axis :
- X
- description :
- Longitude of the center of the grid cell
- long_name :
- longitude
- standard_name :
- longitude
array([235.644501, 235.686157, 235.727829, ..., 245.769333, 245.811005, 245.852661]) - time(time)object2010-01-15 00:00:00 2010-02-15 0...
- description :
- days since 1900-01-01
array([cftime.DatetimeNoLeap(2010, 1, 15, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2010, 2, 15, 0, 0, 0, 0, has_year_zero=True)], dtype=object)
- long_name :
- Monthly Average of Daily Maximum Near-Surface Air Temperature
- units :
- K
- grid_mapping :
- crs
- standard_name :
- air_temperature
- height :
- 2 m
- cell_methods :
- time: maximum(interval: 24 hours);mean over days
- _ChunkSizes :
- [ 10 44 107]
<xarray.DataArray 'air_temperature' (time: 2)> array([286.05795, 286.02655], dtype=float32) Coordinates: * time (time) object 2010-01-15 00:00:00 2010-02-15 00:00:00
- time: 2
- 286.1 286.0
array([286.05795, 286.02655], dtype=float32)
- time(time)object2010-01-15 00:00:00 2010-02-15 0...
- description :
- days since 1900-01-01
array([cftime.DatetimeNoLeap(2010, 1, 15, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2010, 2, 15, 0, 0, 0, 0, has_year_zero=True)], dtype=object)
# Create a 3d mask - this contains the true / false values identifying pixels
# inside vs outside of the mask region
cali_mask = regionmask.mask_3D_geopandas(cali_aoi,
monthly_forecast_temp_xr.lon,
monthly_forecast_temp_xr.lat)
cali_mask
<xarray.DataArray 'region' (region: 1, lat: 585, lon: 1386)>
array([[[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False],
...,
[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False]]])
Coordinates:
* lat (lat) float64 25.06 25.1 25.15 25.19 ... 49.27 49.31 49.35 49.4
* lon (lon) float64 235.2 235.3 235.3 235.4 ... 292.8 292.9 292.9 292.9
* region (region) int64 53- region: 1
- lat: 585
- lon: 1386
- False False False False False False ... False False False False False
array([[[False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], ..., [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False]]]) - lat(lat)float6425.06 25.1 25.15 ... 49.35 49.4
array([25.063078, 25.104744, 25.14641 , ..., 49.312691, 49.354359, 49.396023])
- lon(lon)float64235.2 235.3 235.3 ... 292.9 292.9
array([235.227844, 235.269501, 235.311157, ..., 292.851929, 292.893585, 292.935242]) - region(region)int6453
array([53])
To make processing a bit quicker, below you slice out two months of the data.
# Slice out two months of data
two_months = monthly_forecast_temp_xr.sel(
time=slice('2099-10-25', '2099-12-15'))
Next, apply the mask for just the state of California to the data.
# Apply the mask for California to the data
two_months = two_months.where(cali_mask)
two_months
<xarray.Dataset>
Dimensions: (time: 2, lat: 585, lon: 1386, region: 1, crs: 1)
Coordinates:
* lat (lat) float64 25.06 25.1 25.15 25.19 ... 49.31 49.35 49.4
* crs (crs) int32 1
* lon (lon) float64 235.2 235.3 235.3 235.4 ... 292.9 292.9 292.9
* time (time) object 2099-11-15 00:00:00 2099-12-15 00:00:00
* region (region) int64 53
Data variables:
air_temperature (time, lat, lon, region) float32 nan nan nan ... nan nan
Attributes: (12/46)
description: Multivariate Adaptive Constructed Analog...
id: MACAv2-METDATA
naming_authority: edu.uidaho.reacch
Metadata_Conventions: Unidata Dataset Discovery v1.0
Metadata_Link:
cdm_data_type: FLOAT
... ...
contributor_role: Postdoctoral Fellow
publisher_name: REACCH
publisher_email: reacch@uidaho.edu
publisher_url: http://www.reacchpna.org/
license: Creative Commons CC0 1.0 Universal Dedic...
coordinate_system: WGS84,EPSG:4326- time: 2
- lat: 585
- lon: 1386
- region: 1
- crs: 1
- lat(lat)float6425.06 25.1 25.15 ... 49.35 49.4
- long_name :
- latitude
- standard_name :
- latitude
- units :
- degrees_north
- axis :
- Y
- description :
- Latitude of the center of the grid cell
array([25.063078, 25.104744, 25.14641 , ..., 49.312691, 49.354359, 49.396023])
- crs(crs)int321
- grid_mapping_name :
- latitude_longitude
- longitude_of_prime_meridian :
- 0.0
- semi_major_axis :
- 6378137.0
- inverse_flattening :
- 298.257223563
array([1], dtype=int32)
- lon(lon)float64235.2 235.3 235.3 ... 292.9 292.9
- units :
- degrees_east
- axis :
- X
- description :
- Longitude of the center of the grid cell
- long_name :
- longitude
- standard_name :
- longitude
array([235.227844, 235.269501, 235.311157, ..., 292.851929, 292.893585, 292.935242]) - time(time)object2099-11-15 00:00:00 2099-12-15 0...
- description :
- days since 1900-01-01
array([cftime.DatetimeNoLeap(2099, 11, 15, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2099, 12, 15, 0, 0, 0, 0, has_year_zero=True)], dtype=object) - region(region)int6453
array([53])
- air_temperature(time, lat, lon, region)float32nan nan nan nan ... nan nan nan nan
- long_name :
- Monthly Average of Daily Maximum Near-Surface Air Temperature
- units :
- K
- grid_mapping :
- crs
- standard_name :
- air_temperature
- height :
- 2 m
- cell_methods :
- time: maximum(interval: 24 hours);mean over days
- _ChunkSizes :
- [ 10 44 107]
array([[[[nan], [nan], [nan], ..., [nan], [nan], [nan]], [[nan], [nan], [nan], ..., [nan], [nan], [nan]], [[nan], [nan], [nan], ..., ... ..., [nan], [nan], [nan]], [[nan], [nan], [nan], ..., [nan], [nan], [nan]], [[nan], [nan], [nan], ..., [nan], [nan], [nan]]]], dtype=float32)
- description :
- Multivariate Adaptive Constructed Analogs (MACA) method, version 2.3,Dec 2013.
- id :
- MACAv2-METDATA
- naming_authority :
- edu.uidaho.reacch
- Metadata_Conventions :
- Unidata Dataset Discovery v1.0
- Metadata_Link :
- cdm_data_type :
- FLOAT
- title :
- Monthly aggregation of downscaled daily meteorological data of Monthly Average of Daily Maximum Near-Surface Air Temperature from College of Global Change and Earth System Science, Beijing Normal University (BNU-ESM) using the run r1i1p1 of the rcp45 scenario.
- summary :
- This archive contains monthly downscaled meteorological and hydrological projections for the Conterminous United States at 1/24-deg resolution. These monthly values are obtained by aggregating the daily values obtained from the downscaling using the Multivariate Adaptive Constructed Analogs (MACA, Abatzoglou, 2012) statistical downscaling method with the METDATA (Abatzoglou,2013) training dataset. The downscaled meteorological variables are maximum/minimum temperature(tasmax/tasmin), maximum/minimum relative humidity (rhsmax/rhsmin),precipitation amount(pr), downward shortwave solar radiation(rsds), eastward wind(uas), northward wind(vas), and specific humidity(huss). The downscaling is based on the 365-day model outputs from different global climate models (GCMs) from Phase 5 of the Coupled Model Inter-comparison Project (CMIP3) utlizing the historical (1950-2005) and future RCP4.5/8.5(2006-2099) scenarios.
- keywords :
- monthly, precipitation, maximum temperature, minimum temperature, downward shortwave solar radiation, specific humidity, wind velocity, CMIP5, Gridded Meteorological Data
- keywords_vocabulary :
- standard_name_vocabulary :
- CF-1.0
- history :
- No revisions.
- comment :
- geospatial_bounds :
- POLYGON((-124.7722 25.0631,-124.7722 49.3960, -67.0648 49.3960,-67.0648, 25.0631, -124.7722,25.0631))
- geospatial_lat_min :
- 25.0631
- geospatial_lat_max :
- 49.3960
- geospatial_lon_min :
- -124.7722
- geospatial_lon_max :
- -67.0648
- geospatial_lat_units :
- decimal degrees north
- geospatial_lon_units :
- decimal degrees east
- geospatial_lat_resolution :
- 0.0417
- geospatial_lon_resolution :
- 0.0417
- geospatial_vertical_min :
- 0.0
- geospatial_vertical_max :
- 0.0
- geospatial_vertical_resolution :
- 0.0
- geospatial_vertical_positive :
- up
- time_coverage_start :
- 2091-01-01T00:0
- time_coverage_end :
- 2095-12-31T00:00
- time_coverage_duration :
- P5Y
- time_coverage_resolution :
- P1M
- date_created :
- 2014-05-15
- date_modified :
- 2014-05-15
- date_issued :
- 2014-05-15
- creator_name :
- John Abatzoglou
- creator_url :
- http://maca.northwestknowledge.net
- creator_email :
- jabatzoglou@uidaho.edu
- institution :
- University of Idaho
- processing_level :
- GRID
- project :
- contributor_name :
- Katherine C. Hegewisch
- contributor_role :
- Postdoctoral Fellow
- publisher_name :
- REACCH
- publisher_email :
- reacch@uidaho.edu
- publisher_url :
- http://www.reacchpna.org/
- license :
- Creative Commons CC0 1.0 Universal Dedication(http://creativecommons.org/publicdomain/zero/1.0/legalcode)
- coordinate_system :
- WGS84,EPSG:4326
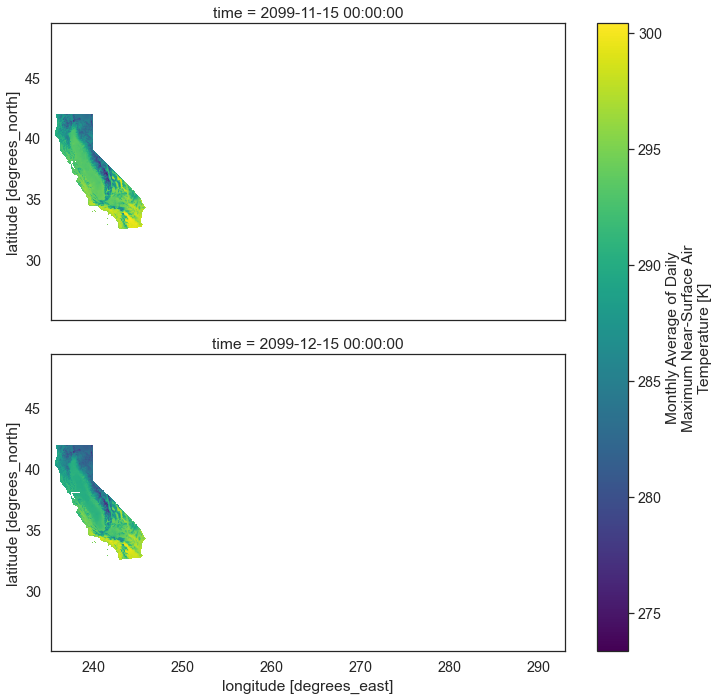
Below you can see that the data for only the region of interest - California, is plotted. However, you have a lot of extra white space surrounding the data.
two_months["air_temperature"].plot(col='time',
col_wrap=1,
figsize=(10, 10))
plt.show()
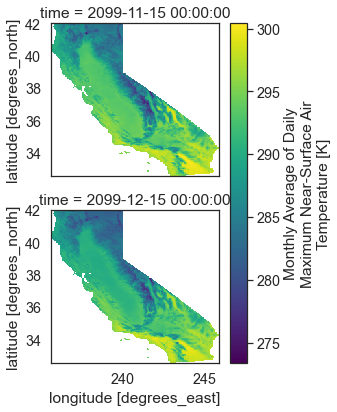
If you slice our your data by time and lat lon and apply the mask for your AOI you get the d
two_months_masked = monthly_forecast_temp_xr["air_temperature"].sel(time=slice('2099-10-25',
'2099-12-15'),
lon=slice(aoi_lon[0],
aoi_lon[1]),
lat=slice(aoi_lat[0],
aoi_lat[1])).where(cali_mask)
two_months_masked.dims
('time', 'lat', 'lon', 'region')
two_months_masked.plot(col='time', col_wrap=1)
plt.show()
Workflow with Multiple Regions
In the example above, you had one region of interest. Below you will implement a workflow where you have multiple regions. For this example, you will subset multiple states from your shapefile.
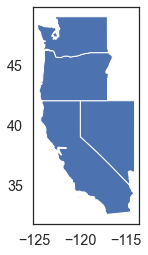
# Start by extracting a few states from the states_gdf
states_gdf["name"]
cali_or_wash_nev = states_gdf[states_gdf.name.isin(
["California", "Oregon", "Washington", "Nevada"])]
cali_or_wash_nev.plot()
plt.show()
Next, create a mask just like you did above. However this time you will use the new extent that contains 4 states (4 regions).
west_mask = regionmask.mask_3D_geopandas(cali_or_wash_nev,
monthly_forecast_temp_xr.lon,
monthly_forecast_temp_xr.lat)
west_mask
<xarray.DataArray 'region' (region: 4, lat: 585, lon: 1386)>
array([[[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False],
...,
[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False]],
[[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False],
...,
[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False]],
[[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False],
...,
[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False]],
[[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False],
...,
[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False]]])
Coordinates:
* lat (lat) float64 25.06 25.1 25.15 25.19 ... 49.27 49.31 49.35 49.4
* lon (lon) float64 235.2 235.3 235.3 235.4 ... 292.8 292.9 292.9 292.9
* region (region) int64 53 82 86 96- region: 4
- lat: 585
- lon: 1386
- False False False False False False ... False False False False False
array([[[False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], ..., [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False]], [[False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], ..., [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False]], [[False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], ..., [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False]], [[False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], ..., [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False], [False, False, False, ..., False, False, False]]]) - lat(lat)float6425.06 25.1 25.15 ... 49.35 49.4
array([25.063078, 25.104744, 25.14641 , ..., 49.312691, 49.354359, 49.396023])
- lon(lon)float64235.2 235.3 235.3 ... 292.9 292.9
array([235.227844, 235.269501, 235.311157, ..., 292.851929, 292.893585, 292.935242]) - region(region)int6453 82 86 96
array([53, 82, 86, 96])
Below is a small helper function that grabs the AOI from a shapefile and returns a dictionary that you can use to slice your data.
def get_aoi(shp, world=True):
"""Takes a geopandas object and converts it to a lat/ lon
extent
Parameters
-----------
shp : GeoPandas GeoDataFrame
A geodataframe containing the spatial boundary of interest
world : boolean
True if you want lat / long to represent sinusoidal (0-360 degrees)
Returns
-------
Dictionary of lat and lon spatial bounds
"""
lon_lat = {}
# Get lat min, max
aoi_lat = [float(shp.total_bounds[1]), float(shp.total_bounds[3])]
aoi_lon = [float(shp.total_bounds[0]), float(shp.total_bounds[2])]
if world:
aoi_lon[0] = aoi_lon[0] + 360
aoi_lon[1] = aoi_lon[1] + 360
lon_lat["lon"] = aoi_lon
lon_lat["lat"] = aoi_lat
return lon_lat
west_bounds = get_aoi(cali_or_wash_nev)
Similar to your workflow above, you can subset that data as you wish by time and extent. Subsetting the data will make all of your data processing faster!
# Slice the data
start_date = "2010-01-15"
end_date = "2010-02-15"
# Subset
two_months_west_coast = monthly_forecast_temp_xr["air_temperature"].sel(
time=slice(start_date, end_date),
lon=slice(west_bounds["lon"][0], west_bounds["lon"][1]),
lat=slice(west_bounds["lat"][0], west_bounds["lat"][1]))
two_months_west_coast
<xarray.DataArray 'air_temperature' (time: 2, lat: 395, lon: 256)>
[202240 values with dtype=float32]
Coordinates:
* lat (lat) float64 32.56 32.6 32.65 32.69 ... 48.85 48.9 48.94 48.98
* lon (lon) float64 235.3 235.4 235.4 235.4 ... 245.8 245.9 245.9 245.9
* time (time) object 2010-01-15 00:00:00 2010-02-15 00:00:00
Attributes:
long_name: Monthly Average of Daily Maximum Near-Surface Air Tempera...
units: K
grid_mapping: crs
standard_name: air_temperature
height: 2 m
cell_methods: time: maximum(interval: 24 hours);mean over days
_ChunkSizes: [ 10 44 107]- time: 2
- lat: 395
- lon: 256
- ...
[202240 values with dtype=float32]
- lat(lat)float6432.56 32.6 32.65 ... 48.94 48.98
- long_name :
- latitude
- standard_name :
- latitude
- units :
- degrees_north
- axis :
- Y
- description :
- Latitude of the center of the grid cell
array([32.562958, 32.604626, 32.64629 , ..., 48.89603 , 48.937698, 48.979362])
- lon(lon)float64235.3 235.4 235.4 ... 245.9 245.9
- units :
- degrees_east
- axis :
- X
- description :
- Longitude of the center of the grid cell
- long_name :
- longitude
- standard_name :
- longitude
array([235.311157, 235.352844, 235.394501, ..., 245.852661, 245.894333, 245.936005]) - time(time)object2010-01-15 00:00:00 2010-02-15 0...
- description :
- days since 1900-01-01
array([cftime.DatetimeNoLeap(2010, 1, 15, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2010, 2, 15, 0, 0, 0, 0, has_year_zero=True)], dtype=object)
- long_name :
- Monthly Average of Daily Maximum Near-Surface Air Temperature
- units :
- K
- grid_mapping :
- crs
- standard_name :
- air_temperature
- height :
- 2 m
- cell_methods :
- time: maximum(interval: 24 hours);mean over days
- _ChunkSizes :
- [ 10 44 107]
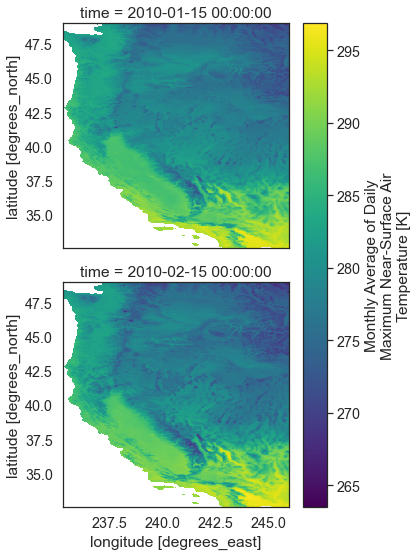
Plot the data.
two_months_west_coast.plot(col="time",
col_wrap=1,
figsize=(5, 8))
plt.show()
Again following the same steps as above, you can apply your spatial mask. This will return a new array with the air temperature data that can be grouped by each of the 4 regions.
# Apply the mask for the west coast subset
two_months_west_coast = two_months_west_coast.where(west_mask)
two_months_west_coast.dims
('time', 'lat', 'lon', 'region')
By default region mask takes the index of the geopandas object and uses it as a region identifier. If you remember from above, california was 53.
cali_or_wash_nev
| featurecla | scalerank | adm1_code | diss_me | iso_3166_2 | wikipedia | iso_a2 | adm0_sr | name | name_alt | ... | name_nl | name_pl | name_pt | name_ru | name_sv | name_tr | name_vi | name_zh | ne_id | geometry | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 53 | Admin-1 scale rank | 2 | USA-3521 | 3521 | US-CA | http://en.wikipedia.org/wiki/California | US | 8 | California | CA|Calif. | ... | Californië | Kalifornia | Califórnia | Калифорния | Kalifornien | Kaliforniya | California | 加利福尼亚州 | 1159308415 | MULTIPOLYGON (((-114.61054 34.99112, -114.6109... |
| 82 | Admin-1 scale rank | 2 | USA-3523 | 3523 | US-NV | http://en.wikipedia.org/wiki/Nevada | US | 1 | Nevada | NV|Nev. | ... | Nevada | Nevada | Nevada | Невада | Nevada | Nevada | Nevada | 内华达州 | 1159315345 | POLYGON ((-114.04027 37.00415, -114.04117 36.6... |
| 86 | Admin-1 scale rank | 2 | USA-3525 | 3525 | US-OR | http://en.wikipedia.org/wiki/Oregon | US | 6 | Oregon | OR|Ore. | ... | Oregon | Oregon | Oregon | Орегон | Oregon | Oregon | Oregon | 俄勒冈州 | 1159309549 | POLYGON ((-120.00081 42.00056, -120.26446 42.0... |
| 96 | Admin-1 scale rank | 2 | USA-3519 | 3519 | US-WA | http://en.wikipedia.org/wiki/Washington_(state) | US | 6 | Washington | WA|Wash. | ... | Washington | Waszyngton | Washington | Вашингтон | Washington | Vaşington | Washington | 华盛顿州 | 1159309547 | MULTIPOLYGON (((-122.78878 48.99303, -122.6863... |
4 rows × 84 columns
# Note that the region values align with the index of the geodataframe above
two_months_west_coast.region
<xarray.DataArray 'region' (region: 4)> array([53, 82, 86, 96]) Coordinates: * region (region) int64 53 82 86 96
- region: 4
- 53 82 86 96
array([53, 82, 86, 96])
- region(region)int6453 82 86 96
array([53, 82, 86, 96])
Having an additional region dimension in your array makes it easier to summarize your data by region. Below you plot the data by time and region in a matrix.
two_months_west_coast.plot(col="time",
row="region",
sharey=False, sharex=False)
plt.show()

Calculate Mean Air Temperature by Region
Once you have your data subsetted by region, you can calculate summary statistics on the data for each region.
Below, you calculate the mean air temperature value for the time subset that you created above. At this point you could export the dataframe to a csv file if you want!
summary = two_months_west_coast.groupby("time").mean(["lat", "lon"])
summary.to_dataframe()
| air_temperature | ||
|---|---|---|
| time | region | |
| 2010-01-15 00:00:00 | 53 | 286.057922 |
| 82 | 280.085175 | |
| 86 | 279.390350 | |
| 96 | 278.031158 | |
| 2010-02-15 00:00:00 | 53 | 286.026550 |
| 82 | 279.355438 | |
| 86 | 278.442688 | |
| 96 | 277.346008 |
The Entire Workflow To Subset netcdf 4 Files by Temporal and Spatial Extents
Putting all of the above together, below is an example workflow that you might use to summarize climate data in netcdf 4 format by region and over time.
# Helper Function to extract AOI
def get_aoi(shp, world=True):
"""Takes a geopandas object and converts it to a lat/ lon
extent """
lon_lat = {}
# Get lat min, max
aoi_lat = [float(shp.total_bounds[1]), float(shp.total_bounds[3])]
aoi_lon = [float(shp.total_bounds[0]), float(shp.total_bounds[2])]
# Handle the 0-360 lon values
if world:
aoi_lon[0] = aoi_lon[0] + 360
aoi_lon[1] = aoi_lon[1] + 360
lon_lat["lon"] = aoi_lon
lon_lat["lat"] = aoi_lat
return lon_lat
# Here is the entire workflow from start to end
url = (
"https://naturalearth.s3.amazonaws.com/"
"50m_cultural/ne_50m_admin_1_states_provinces_lakes.zip"
)
states_gdf = gpd.read_file(url)
# Get netcdf file
data_path_monthly = 'http://thredds.northwestknowledge.net:8080/thredds/dodsC/agg_macav2metdata_tasmax_BNU-ESM_r1i1p1_rcp45_2006_2099_CONUS_monthly.nc'
# Open up the data
with xr.open_dataset(data_path_monthly) as file_nc:
monthly_forecast_temp_xr = file_nc
# View xarray object
monthly_forecast_temp_xr
# Start by extracting a few states from the states_gdf
# I am using this as a shapefile because i presume some of my students
# will want to use custom aoi's so i want to teach using a shapefil rather than
# the regionmask built in shapes as nice as they are :)
states_gdf["name"]
cali_or_wash_nev = states_gdf[states_gdf.name.isin(
["California", "Oregon", "Washington", "Nevada"])]
west_mask = regionmask.mask_3D_geopandas(cali_or_wash_nev,
monthly_forecast_temp_xr.lon,
monthly_forecast_temp_xr.lat)
west_mask
west_bounds = get_aoi(cali_or_wash_nev)
# Slice the data
start_date = "2010-01-15"
end_date = "2020-02-15"
# Subset the data - this is now a dataarray rather than a DataSet
two_months_west_coast = monthly_forecast_temp_xr["air_temperature"].sel(
time=slice(start_date, end_date),
lon=slice(west_bounds["lon"][0], west_bounds["lon"][1]),
lat=slice(west_bounds["lat"][0], west_bounds["lat"][1])).where(west_mask)
# This output is a dataarray
two_months_west_coast
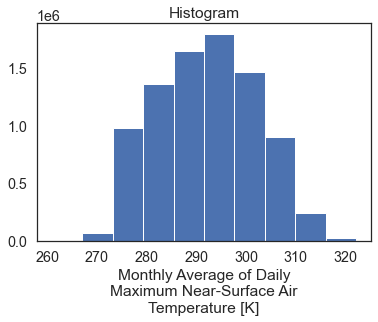
# Plot the data - this produces a single histogram
two_months_west_coast.plot()
plt.show()
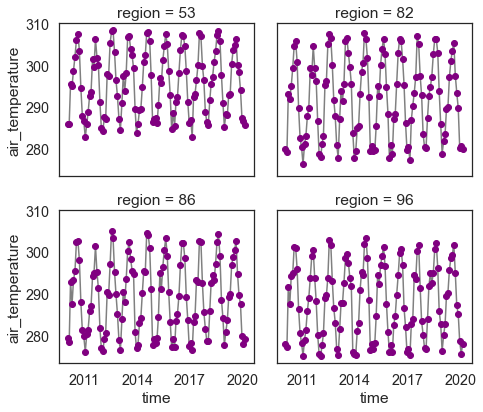
Once the data are subsetted, you can begin to create plots and dataframes with the summary data.
# Keep in mind that as you add more data to your slice, the processing will slow down.
regional_summary = two_months_west_coast.groupby("region").mean(["lat", "lon"])
regional_summary.plot(col="region",
marker="o",
color="grey",
markerfacecolor="purple",
markeredgecolor="purple",
col_wrap=2)
plt.show()
Export to a dataframe.
two_months_west_coast.groupby("region").mean(["lat", "lon"]).to_dataframe()
| air_temperature | ||
|---|---|---|
| time | region | |
| 2010-01-15 00:00:00 | 53 | 286.057922 |
| 82 | 280.085175 | |
| 86 | 279.390350 | |
| 96 | 278.031158 | |
| 2010-02-15 00:00:00 | 53 | 286.026550 |
| ... | ... | ... |
| 2020-01-15 00:00:00 | 96 | 275.706940 |
| 2020-02-15 00:00:00 | 53 | 285.709076 |
| 82 | 279.856842 | |
| 86 | 279.198822 | |
| 96 | 278.026459 |
488 rows × 1 columns
Share on
Twitter Facebook Google+ LinkedIn
Leave a Comment