Lesson 3. Use tidyverse group_by and summarise to Manipulate Data in R Clean coding tidyverse intro
Learning objectives
At the end of this activity, you will be able to:
- Use the
group_by,summariseandmutatefunctions to manipulate data inR. - Use
readrto open tabular data inR. - Read CSV data files by specifying a URL in
R. - Work with no data values in
R.
What you need
We recommend that you have R and RStudio setup to complete this lesson. You will also need the following R packages:
- ggplot2
- dplyr
- readr
- lubridate
About the Data
The data that you will use for this workshop is stored in the cloud. It contains precipitation information over time for several locations in Colorado.
All you have to get started with is a list of URLs - one for each data file. Each data file is in .csv format. You can find this list of URLs in the data/ directory of the version-control-hot-mess GitHub repository that you cloned or downloaded for this workshop.
Data Exploration
To begin this lesson you will explore your data.
What Is the Length of Record For Each Site?
Your end goal in this workshop is to create plots of precipitation data over time by station and month / year. However, you have yet to explore your data. To begin, open the first url in csv file containing urls of the data locations. Remember that file is located in data/data_urls.csv.
Explore your data and calculate the length of record for each site in the data.
For this activity you will use the readr library to import your data - a powerful library for parsing and reading tabular data. The readr package will attempt to convert known character formats including date/times, numbers and other formats into the correct R class.
# load libraries
library(readr)
library(ggplot2)
library(dplyr)
Next, open the file that contains URLs to the data. Note that we are using data that are stored on Amazon Web Services (AWS) servers.
# import data using readr
all_paths <- read_csv("data/data_urls.csv")
glimpse(all_paths)
## Observations: 33
## Variables: 1
## $ url <chr> "https://s3-us-west-2.amazonaws.com/earthlab-teaching/vchm/M…
Open a File with readr::read_csv
Next, open the data contained in the first URL in the .csv file that you just imported above.
# grab first URL from the file
first_csv <- all_paths$url[1]
# open the first data file using readr:read_csv
year_one <- read_csv(first_csv)
Note that when you use readr::read_csv, it returns the data class that each column was converted to. Above, notice that the lat, lon, elevation are all of type double - which is a number with decimal places.
The DATE field was converted to a proper datetime class.
The HPCP column stores precipitation. This is the data that you ultimately want to plot. Notice that those data were not converted to a numeric format. You will explore that issue later in this lesson.
What is Pseudocode?
Before you start to code, think about your goals. Rather than simply jumping into R and coding (which is what we all want to do initially!), plan things out.
Write down that steps associated with what you wish to accomplish - in English. Writing out the steps required to complete an operation is called pseudocode. Pseudocode is useful for organization coding operations. It allows you to think through what you wish to accomplish and the most efficient way to go about it BEFORE you write your code.
GOAL: You want to calculate the total time in days that is represented in the precipitation data for colorado for each station or site.
Write Pseudocode
Once your goal is clear, write out the steps that you will need to implement in order to achieve your goal. It’s ok if you don’t know all of the functions yet to implement this. Organize first, look up functions second.
## Below is the pseudocode for calculating length of record
# 1. open up the file containing the data
# 2. group by data by the station name field
# 3. calculate the total time by subtracting the min date from the max date.
Once your pseudocode is written out, it’s time to associated R functions with each step. To do that you will use the tidyverse.
Get Started with tidyverse
To get going with tidyverse, there are a few things that you should know.
- The pipe
%>%is fundamental to tidyverse. The pipe is a way to connect a sequence of operations together. Pipes are efficient because they:
- Don’t create intermediate outputs saving memory
- Combine operations into a clean chunk of code
- Allow you to send one output as an input to the next operation.
When combined with tidyverse functions, you also gain extremely expressive code. Pipes generally are often used with a data.frame object and are written as follows:
my_data_frame %>%
perform_some_operation
Pipes are a powerful tool for clearly expressing a sequence of multiple operations. - Hadley Wickham, R for Data Science
R tidyverse summarise and group_by Functions
The next operations that you need to know are the summarise and group_by functions.
group_by: As the name suggest,group_byallows you to group by a one or more variables.summarize:summarizecreates a newdata.framecontaining calculated summary information about a grouped variable.
group_by and summarize are two of the most commonly used tidyverse functions. For example:
# group_by / summarise workflow example
my_data_frame %>%
group_by(total_precip_col) %>%
summarise(avg_precip = mean(total_precip_col))
Calculate Total Days of Observations
You can calculate the total number of days represented in your data by subtracting the maximum date from the minimun date for each station. The dates were stored in a friendly format that readr could understand and convert to a datetime class.
Your code to calculate length of record will thus look something like this:
# 1. open up the file containing the data
read_csv(first_csv) %>%
# 2. group by data by the station name field
group_by(STATION_NAME) %>%
# 3. calculate the total time by subtracting the min date from the max date.
summarize(total_days = max(DATE) - min(DATE))
## # A tibble: 4 x 2
## STATION_NAME total_days
## <chr> <drtn>
## 1 BOULdER 2 CO US 0.0000 secs
## 2 BOULDEr 2 CO US 0.0000 secs
## 3 BOULDER 2 cO US 113.5417 secs
## 4 BOULDER 2 CO US 334.6250 secs
On Your Own (OYO)
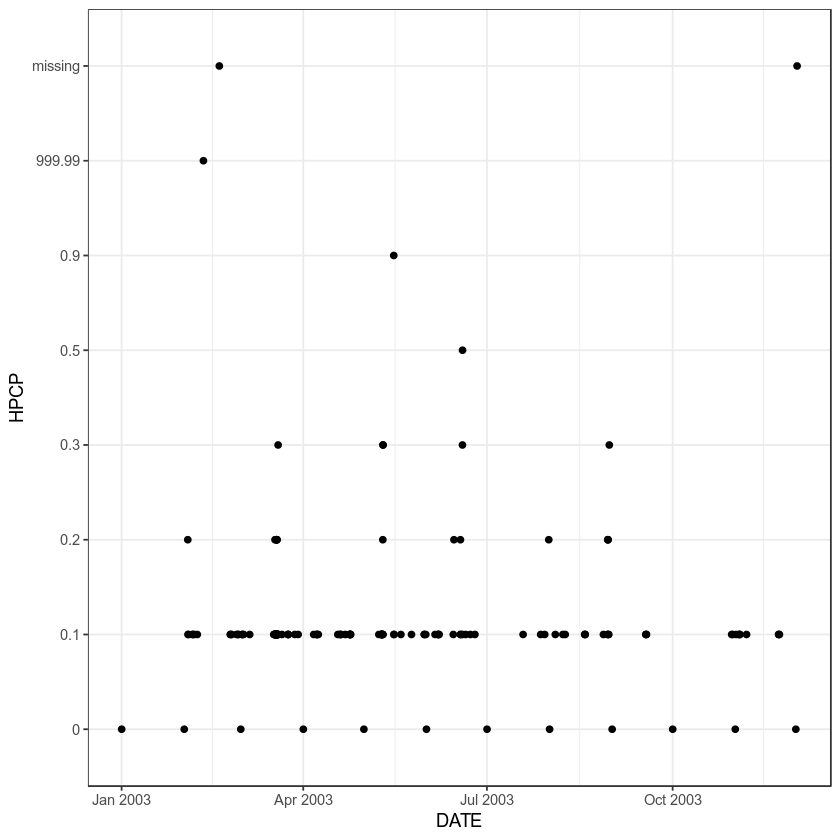
Create a plot of precipitation over time using the .csv file that is accessed through the first URL in the list. This is the same file we’ve been using throughout this lesson. To help you create your plot, an example of creating a scatter plot with ggplot and sending a data.frame to ggplot is below.
# Syntax to create scatter plot using ggplot
data.frame %>%
ggplot(aes(x = date_field_here, y = precipitation_field_here)) +
geom_point() +
theme_bw()
Note that the code above does NOT create the plot below! It provides you with the syntax that you need to create the plot.
Additional resources
You may find the materials below useful as an overview of what we cover during this workshop:
Share on
Twitter Facebook Google+ LinkedIn
Leave a Comment