Lesson 2. Get Started with Clean Coding in R Clean coding tidyverse intro
Learning objectives
At the end of this activity, you will be able to:
- Identify issues that can be improved in your code in the areas of syntax, modularity, documentation and expressiveness.
- Define syntax, documentation, modularity and expressiveness.
What you need
- You need internet access to complete this activity
What is Clean Code?
The theme of this workshop is clean code. When we say clean code, we are referring to code that is written in a way that is easy to understand (for you, your future self and your colleagues), efficient in it’s implementation and well documented.
In this workshop, we will explore several tricks and tips that you can use to write more efficient code. These include:
- Writing pseudocode before executable code to organize your approach. Pseudocode refers to writing out the steps and process that you will need to implement in your code - in plain old english - BEFORE you code. We will discuss pseudocode more in the next lesson.
- Writing more expressive code by using meaningful variable names and
tidyversefunctions and syntax inR. - Automating your code rather than using a “copy pasta” approach. Writing pseudocode can help with identifying repetitive tasks.
Core Principles of Clean Code
In this workshop, you will learn how to write better, more efficient and easier to read, well-documented code. You can segment the concept of clean code into 4 main components:
- Syntax: Syntax is the format or style that you use to write your code. When a group of scientists use the same syntax, the code becomes more familiar and thus easier to quicly scan, read and understnd. Different style guides have been developed
- Modularity: Is your code more “script like” written from beginning to end with repeated tasks? Or does it consist of sections and functions that capture tasks that are repeated?
- Documentation: Is your code well documented? Documentation can range from comments found within your code to thorough
readmefiles that describe your entire workflow. - Expressiveness: Expressiveness refers to writing code that makes your intention transparent. This includes using meaningful names for objects and files, that indicate something about their contents and intended use.
Don’t Repeat Yourself (DRY)
The DRY approach to programming refers to writing functions and automating sections of code that are repeated. If you perform the same task multiple times in your code, consider a function or a loop to make your workflow more efficient.
About The Data
The goal of this workshop is as follows:
You have been given access to some precipitation data for Colorado across several locations This data is stored in the cloud and your colleague has started to explore the data in R.
Your colleague has given you two things:
- A
.csvfile containing a list of URL’s for each.csvdata file - Some thoughtfully composed code (see below) that they wrote to explore the data and help you get started.
Unfortunately, your colleague is trekking among the tallest peaks in the Himalaya. Thus, you are left to your own crafty devices to figure out how to work with these data.
Your goal in this workshop is to create a plot of the data in R. Something like this:
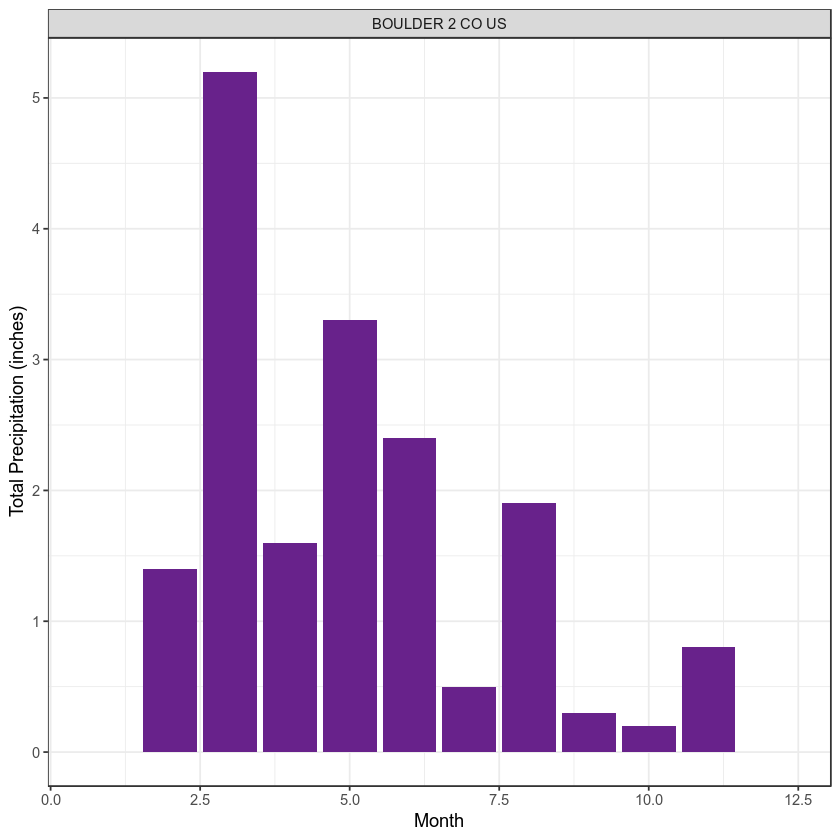
Time to get started. Let’s begin by having a close look at the thoughtful code that your colleague left for you to work with.
Activity One
You are working with some time series data that captures precipitation for several study sites in Colorado. A colleague of yours provides you with the code below that they started to develop to work with the data.
Do the following:
- Break up into groups of 2-3.
- Identify any issues that you find with that code.
- Add the issues that you find in the code to the Google Doc provided for the workshop. In some cases, someone else may have already identified and issue that you did. Add a +1 to any items that you agree with so issues are duplicated. Also feel free to edit / add to any issues you see in the document that you’d like to build upon.
Be sure to identify the issues as they relate to one (or more) of the four categories that we defined above
- Syntax,
- Modularity
- Documentation
- Expressiveness
You have about 15 minutes to complete this task! We will discuss what everyone finds as a group.
# cause ggplot is the coolest
library(ggplot)
#### Commence analysis
myDATA <- read.csv("https://s3-us-west-2.amazonaws.com/earthlab-teaching/vchm/My_Data2003.csv")
#uh oh -- why are things messed up?
head(myDATA)
# i really want to create a nice dataframe that summarizes rainfall by month in mm.
# so let's see how this goes.
#year
# First I need to make sure that the DATE column is a Date!!!
myDATA$dateasdate <- as.Date(myDATA$DATE)
str(myDATA) # check to make sure that the date column is a date - is it a date?
# Then I want to make sure that I have a column that is the month and another one that is year
myDATA$Month <- lubridate::month(myDATA$dateasdate)
str(myDATA) # check to make sure that the date column is a date
# Then I want to have a YEAR column that is the YEAR
myDATA$year <- lubridate::year(myDATA$dateasdate)
str(myDATA) # check to make sure that the date column is a date
#myDATA$DOY <- as.numeric(strftime(myDATA$dateasdate, format = "%j"))
#
# library(stringr)
# a_year <- myDATA %>%
# mutate(DATE = gsub("/", "-", DATE),
# DATE = ) %>%
# mutate(DATE2 = as.POSIXct(DATE, format = "%d-%m-%y %H:%M"),
# DATE3 = as.POSIXct(DATE, format = "%d-%m-%Y %H:%M"))
# a_year$DATE2 <- gsub("(.*)-(..)$", "\\1-20\\2", a_year$DATE)
# fix the year with regex
# gsub("(.*)-(..)$", "\\1-20\\2", vec1)
#
# gsub("(.*)/(..)$", "\\1/19\\2", vec1)
# # the dates are all messed up. who made this data? seriously?
# myDATA$DATE
# i don't think this worked but who knows
# Finally, I want to save a CSV file has precip in mm
d2003 <- myDATA[myDATA[[12]] == 2003]
d2003$precip_MM = myDATA$HPCP * 25.4
write.csv(file = 'data/outputs/precip_2003.csv', x = d2003, row.names = FALSE)
as_2003 <- a_year %>%
mutate(precip_mm = HPCP * 25.4,
month = month(DATE))
write.csv(as_2003,"data/outputs/precip_2003.csv")
write.csv(a_year, file = "data/annual/precip-2003.csv")
#year
a_year <- boulder_precip %>%
filter(year(DATE) == 2004) %>%
mutate(month = month(DATE))
write.csv(a_year, file = "data/annual/precip-2003.csv")
library(ggplot2)
#year
a_year <- boulder_precip %>%
filter(year(DATE) == 2005) %>%
mutate(month = month(DATE))
write.csv(a_year, file = "data/annual/precip-2003.csv")
#year
a_year <- boulder_precip %>%
filter(year(DATE) == 2005) %>%
mutate(month = month(DATE))
write.csv(a_year, file = "data/annual/precip-2006.csv")
#year
a_year <- boulder_precip %>%
filter(year(DATE) == 2003) %>%
mutate(month = month(DATE))
write.csv(a_year, file = "data/annual/precip-2007.csv")
#year
a_year <- boulder_precip %>%
filter(year(DATE) == 2003) %>%
mutate(month = month(DATE))
write.csv(a_year, file = "data/annual/precip-2008.csv")
#year
# I don't think that worked we will do it again but without error
#==============================================================================
#==============================================================================
#STRATEGY: going to calculate monthly averages of precipitation for each dataset
# (each year!) using R
#==============================================================================
#==============================================================================
myDATA <- read.csv("https://s3-us-west-2.amazonaws.com/earthlab-teaching/vchm/My_Data2003.csv")
myFinalData <- myDATA
unique(myFinalData$HPCP)
#na values
myDATA <- read.csv("https://s3-us-west-2.amazonaws.com/earthlab-teaching/vchm/My_Data2003.csv",
na.strings = c("999.99"))
myFinalData <- myDATA
unique(myFinalData$HPCP)
myFinalData$DATE2 = as.POSIXct(myFinalData$DATE, format = "%Y-%d-%m %H:%M")
myFinalData$year <- substr(myFinalData$DATE, 1, 4)
myFinalData$month <- substr(myFinalData$DATE, 6, 7)
myFinalData = myFinalData[myFinalData$year == "2003",]
#myFinalData$month <- month(myFinalData$DATE)
#summary mean dataframe 2003
# this is easy to do.
finalSUMMARYmean <- data.frame(jan_mean_2003 = mean(myFinalData$HPCP[myFinalData$month == "01"], na.rm = TRUE),
feb_mean_2003 = mean(myFinalData$HPCP[myFinalData$month == "02"], na.rm = TRUE),
march_mean_2003 = mean(myFinalData$HPCP[myFinalData$month == "03"], na.rm = TRUE),
apr_mean_2003 = mean(myFinalData$HPCP[myFinalData$month == "04"], na.rm = TRUE),
may_mean_2003 = mean(myFinalData$HPCP[myFinalData$month == "05"], na.rm = TRUE),
june_mean_2003 = mean(myFinalData$HPCP[myFinalData$month == "05"], na.rm = TRUE),
may_mean_2003 = mean(myFinalData$HPCP[myFinalData$month == "06"], na.rm = TRUE),
july_mean_2003 = mean(myFinalData$HPCP[myFinalData$month == "07"], na.rm = TRUE),
aug_mean_2003 = mean(myFinalData$HPCP[myFinalData$month == "08"], na.rm = TRUE),
sept_mean_2003 = mean(myFinalData$HPCP[myFinalData$month == "09"], na.rm = TRUE),
oct_mean_2003 = mean(myFinalData$HPCP[myFinalData$month == "09"], na.rm = TRUE),
nov_mean_2003 = mean(myFinalData$HPCP[myFinalData$month == "11"], na.rm = TRUE),
dec_mean_2003 = mean(myFinalData$HPCP[myFinalData$month == "12"], na.rm = TRUE))
finalSUMMARYmean
### 2004 - easy enough
#i'd like a snack
#na values
myDATA <- read.csv("https://s3-us-west-2.amazonaws.com/earthlab-teaching/vchm/My_Data2004.csv",
na.strings = c("999.99"))
unique(myFinalData$HPCP)
myDATA <- read.csv("https://s3-us-west-2.amazonaws.com/earthlab-teaching/vchm/My_Data2004.csv",
na.strings = c(" ", "999.99", "missing", "n/a"))
unique(myDATA$HPCP)
myDATA$DATE2 = as.POSIXct(myDATA$DATE, format = "%Y-%d-%m %H:%M")
myDATA$year <- substr(myDATA$DATE, 1, 4)
myDATA$month <- substr(myDATA$DATE, 6, 7)
myDATA = myDATA[myDATA$year == "2004",]
jan_mean_2003 = mean(myFinalData$HPCP[myFinalData$month == "01"], na.rm = TRUE)
#myFinalData$month <- month(myFinalData$DATE)
#summary mean dataframe 2003
finalSUMMARYmean$jan_mean_2004 <- mean(myDATA$HPCP[myFinalData$month == "01"], na.rm = TRUE)
finalSUMMARYmean$feb_mean_2004 <- mean(myDATA$HPCP[myFinalData$month == "02"], na.rm = TRUE)
finalSUMMARYmean$march_mean_2004 <-mean(myDATA$HPCP[myFinalData$month == "03"], na.rm = TRUE)
finalSUMMARYmean$apr_mean_2004 = mean(myDATA$HPCP[myFinalData$month == "04"], na.rm = TRUE)
finalSUMMARYmean$may_mean_2004 = mean(myDATA$HPCP[myFinalData$month == "05"], na.rm = TRUE)
finalSUMMARYmean$june_mean_2004 = mean(myDATA$HPCP[myFinalData$month == "05"], na.rm = TRUE)
finalSUMMARYmean$may_mean_2004 = mean(myDATA$HPCP[myFinalData$month == "06"], na.rm = TRUE)
finalSUMMARYmean$july_mean_2004 = mean(myDATA$HPCP[myFinalData$month == "07"], na.rm = TRUE)
finalSUMMARYmean$aug_mean_2004 = mean(myDATA$HPCP[myFinalData$month == "08"], na.rm = TRUE)
finalSUMMARYmean$sept_mean_2004 = mean(myDATA$HPCP[myFinalData$month == "09"], na.rm = TRUE)
finalSUMMARYmean$oct_mean_2004 = mean(myDATA$HPCP[myFinalData$month == "09"], na.rm = TRUE)
finalSUMMARYmean$nov_mean_2004 = mean(myDATA$HPCP[myFinalData$month == "11"], na.rm = TRUE)
finalSUMMARYmean$dec_mean_2004 = mean(myDATA$HPCP[myFinalData$month == "12"], na.rm = TRUE)
finalSUMMARYmean
#na values
#2005 - my fingers are tired. may start lifting finger weights to help with endurance.
myDATA <- read.csv("https://s3-us-west-2.amazonaws.com/earthlab-teaching/vchm/My_Data2005.csv",
na.strings = c("999.99"))
unique(myFinalData$HPCP)
myDATA <- read.csv("https://s3-us-west-2.amazonaws.com/earthlab-teaching/vchm/My_Data2005.csv",
na.strings = c(" ", "999.99", "missing", "n/a"))
unique(myDATA$HPCP)
myDATA$DATE2 = as.POSIXct(myDATA$DATE, format = "%Y-%m-%d %H:%M")
myDATA$year <- substr(myDATA$DATE, 1, 4)
myDATA$month <- substr(myDATA$DATE, 6, 7)
myDATA = myDATA[myDATA$year == "2005",]
#myFinalData$month <- month(myFinalData$DATE)
#summary mean dataframe 2003
finalSUMMARYmean$jan_mean_2005 <- mean(myDATA$HPCP[myFinalData$month == "01"], na.rm = TRUE)
finalSUMMARYmean$feb_mean_2005 <- mean(myDATA$HPCP[myFinalData$month == "02"], na.rm = TRUE)
finalSUMMARYmean$march_mean_2005 <-mean(myDATA$HPCP[myFinalData$month == "03"], na.rm = TRUE)
finalSUMMARYmean$apr_mean_2005 = mean(myDATA$HPCP[myFinalData$month == "04"], na.rm = TRUE)
finalSUMMARYmean$may_mean_2005 = mean(myDATA$HPCP[myFinalData$month == "05"], na.rm = TRUE)
finalSUMMARYmean$june_mean_2005 = mean(myDATA$HPCP[myFinalData$month == "05"], na.rm = TRUE)
finalSUMMARYmean$may_mean_2005 = mean(myDATA$HPCP[myFinalData$month == "06"], na.rm = TRUE)
finalSUMMARYmean$july_mean_2005 = mean(myDATA$HPCP[myFinalData$month == "07"], na.rm = TRUE)
finalSUMMARYmean$aug_mean_2005 = mean(myDATA$HPCP[myFinalData$month == "08"], na.rm = TRUE)
finalSUMMARYmean$sept_mean_2005 = mean(myDATA$HPCP[myFinalData$month == "09"], na.rm = TRUE)
finalSUMMARYmean$oct_mean_2005 = mean(myDATA$HPCP[myFinalData$month == "09"], na.rm = TRUE)
finalSUMMARYmean$nov_mean_2005 = mean(myDATA$HPCP[myFinalData$month == "11"], na.rm = TRUE)
finalSUMMARYmean$dec_mean_2005 = mean(myDATA$HPCP[myFinalData$month == "12"], na.rm = TRUE)
finalSUMMARYmean#mostly working I guess will fix later
#na values
#2006 - i need a nap and some cookies. preferably oreos altho vanilla wafers could be nice
myDATA <- read.csv("https://s3-us-west-2.amazonaws.com/earthlab-teaching/vchm/My_Data2006.csv",
na.strings = c("999.99"))
unique(myFinalData$HPCP)
myDATA <- read.csv("https://s3-us-west-2.amazonaws.com/earthlab-teaching/vchm/My_Data2006.csv",
na.strings = c(" ", "999.99", "missing", "n/a"))
unique(myDATA$HPCP)
myDATA$DATE2 = as.POSIXct(myDATA$DATE, format = "%Y-%m-%d %H:%M")
myDATA$year <- substr(myDATA$DATE, 1, 4)
myDATA$month <- substr(myDATA$DATE, 6, 7)
myDATA = myDATA[myDATA$year == "2006",]
#myFinalData$month <- month(myFinalData$DATE)
#summary mean dataframe 2003
finalSUMMARYmean$jan_mean_2006 <- mean(myDATA$HPCP[myFinalData$month == "01"], na.rm = TRUE)
finalSUMMARYmean$feb_mean_2006 <- mean(myDATA$HPCP[myFinalData$month == "02"], na.rm = TRUE)
finalSUMMARYmean$march_mean_2006 <-mean(myDATA$HPCP[myFinalData$month == "03"], na.rm = TRUE)
finalSUMMARYmean$apr_mean_2006 = mean(myDATA$HPCP[myFinalData$month == "04"], na.rm = TRUE)
finalSUMMARYmean$may_mean_2006 = mean(myDATA$HPCP[myFinalData$month == "05"], na.rm = TRUE)
finalSUMMARYmean$june_mean_2006 = mean(myDATA$HPCP[myFinalData$month == "05"], na.rm = TRUE)
finalSUMMARYmean$may_mean_2006 = mean(myDATA$HPCP[myFinalData$month == "06"], na.rm = TRUE)
finalSUMMARYmean$july_mean_2006 = mean(myDATA$HPCP[myFinalData$month == "07"], na.rm = TRUE)
finalSUMMARYmean$aug_mean_2006 = mean(myDATA$HPCP[myFinalData$month == "08"], na.rm = TRUE)
finalSUMMARYmean$sept_mean_2006 = mean(myDATA$HPCP[myFinalData$month == "09"], na.rm = TRUE)
finalSUMMARYmean$oct_mean_2006 = mean(myDATA$HPCP[myFinalData$month == "09"], na.rm = TRUE)
finalSUMMARYmean$nov_mean_2006 = mean(myDATA$HPCP[myFinalData$month == "11"], na.rm = TRUE)
finalSUMMARYmean$dec_mean_2006 = mean(myDATA$HPCP[myFinalData$month == "12"], na.rm = TRUE)
finalSUMMARYmean# Workikng better now!
write.csv(a_year, file = "finalSUMMARYmean.csv")
#start my analysis
# =============================================================================
# FIGURES FOR THE VISUALIZATION OF PRECIPITATION IN BOULDER COUNTY FROM THE
# YEARS 2003 THROUGH 2010 - WE ARE GOING TO PLOT THE MONTHLY MEANS OVER TIME
# TO SEE WHETHER THERE ARE LONG TERM CHANGES IN PRECIPITATION ALL OF THE PLOTS
# SHOULD BE ON THE SAME PANEL - I ONLY WANT TO COLOR THE LINES TO INDICATE
# YEAR AND THE X AXIS IS MONTH AND THE Y AXIS IS AVERAGE WATER
# =============================================================================
# LAST MODIFIED ON NOVEMBER 9 2017 BY MAX JOSEPH
# PRETTY SURE THIS WORKS BUT NO GUARANTEES
read.csv("data/annual/precip-2003.csv")
plot(data)
mydata <- read.csv("data/week_06/outputs/precip_mm/precip-2003.csv")
myData$newcolumn <- mydata$precip * 12 / 2
my.data <-read.csv("data/week_06/outputs/precip_mm/precip-2004.csv")
my.data$newcolumn <- mydata$precip * 12 / 2
my.data<-read.csv("data/week_06/outputs/precip_mm/precip-2006.csv")
my.data$newcolumn <- mydata$precip * 12 / 2
my.data <-read.csv("data/week_06/outputs/precip_mm/precip-2007.csv")
my.data$newcolumn <- mydata$precip * 12 / 2
my.data <-read.csv("data/week_06/outputs/precip_mm/precip-20010.csv")
my.data$newcolumn <- mydata$precip * 12 / 2
lines(mydata)
#export stuff - i should do this for all of my csv files
write.csv(mydata)
my.data <-read.csv("data/week_06/outputs/precip_mm/precip-2008.csv")
my.data$newcolumn <- mydata$month
#testing some stuff with pipes
outputs <- my.data %>%
mutate(newcolumn = "fredyymack") %>%
group_by(month) %>%
summarise(mean(Precip_MM))
plot(mydata)
#export stuff
write.csv(mydata)
my.data <-read.csv("data/week_06/outputs/precip_mm/precip-2009.csv")
my.data$newcolumn <- mydata$month
#testing some stuff with pipes
outputs <- my.data %>%
mutate(newcolumn = "fredyymack") %>% #i dont think i need this column do i?
group_by(month) %>%
summarise(mean(Precip_MM))
plot(mydata)
#export stuff
write.csv(mydata)
my.data <-read.csv("data/week_06/outputs/precip_mm/precip-2006.csv")
my.data$newcolumn <- mydata$month
#testing some stuff with pipes
outputs2 <- my.data %>%
mutate(newcolumn = "fredyymack") %>%
group_by(month) %>%
summarise(mean(Precip_MM))
plot(mydata)
#export stuff
write.csv(mydata)
my.data <-read.csv("data/week_06/outputs/precip_mm/precip-2009.csv")
my.data<-read.csv("data/week_06/outputs/precip_mm/precip-2012.csv")
my.data <-read.csv("data/week_06/outputs/precip_mm/precip-2013.csv")
#not sure why everything isn't plotting
plot(my.data)
plot(my.data)
plot(mydata)
plot(my.data)
plot(my.data)
#testing some stuff with pipes
my.data %>%
mutate(newcolumn = "fredyymack") %>%
group_by(month) %>%
summarise(mean(Precip_MM))
#testing some stuff with pipes
outputs <- my.data %>%
mutate(newcolumn = "fredyymack") %>%
group_by(month) %>%
summarise(mean(Precip_MM))
ggplot(outputs, aes(y=month, x=precipmm)) +
geom_point()
#need to do this to all of my data
#testing some stuff with pipes
#mydata <- my.data %>%
# mutate(newcolumn = "fredyymack") %>%
# group_by(month) %>%
# summarise(mean(Precip_MM))
#export the data to a csv
write.csv(mydata)
library(dplyr)
library(ggplot2)
Additional resources
You may find the materials below useful as an overview of what we cover during this workshop:
Share on
Twitter Facebook Google+ LinkedIn
Leave a Comment